Gene Essentiality & Genetic Interaction Lab
A systems biology lab at the interface of computational and experimental biology
Traver Hart, PhD
We decipher the complex web of relationships that governs how eukaryotic cells process information, respond to their environments, and transform into cancer. As part of The University of Texas MD Anderson Cancer Center, we search for emergent vulnerabilities of tumor cells and develop computational and experimental tools for CRISPR-mediated genetic engineering in mammalian cells.
Research
Functional Genomics and Genetic Interactions
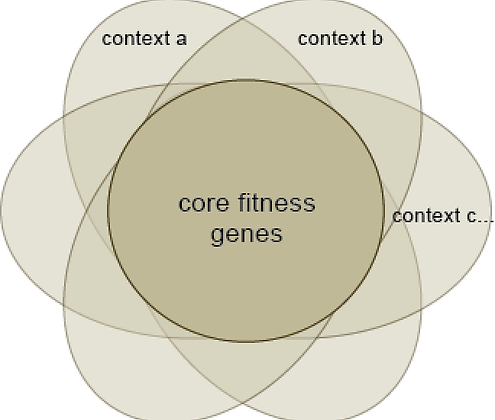
Core vs Context-Specific Essentiality
Cells from different tissues have different genetic dependencies, but all share a common set of "core fitness genes" that are required for proliferation. We use this difference between core and context essentials to guide our estimates of screen quality and gene classification, and to identify tumor-specific pathways and targets.
New CRISPR technologies enable precise multiplex genetic perturbations, allowing us to go beyond simple single-gene knockouts. We can now begin to explore genetic interaction networks in mammalian cells and decipher cellular organization as well as discover synthetic lethals.
Functional Interaction Networks
Genes with correlated knock-out fitness profiles across a diverse set of screens operate in the same biological process or biochemical pathway. But there are many ways to construct these functional interaction networks. Which is best?
Read the paper (Gheorghe & Hart)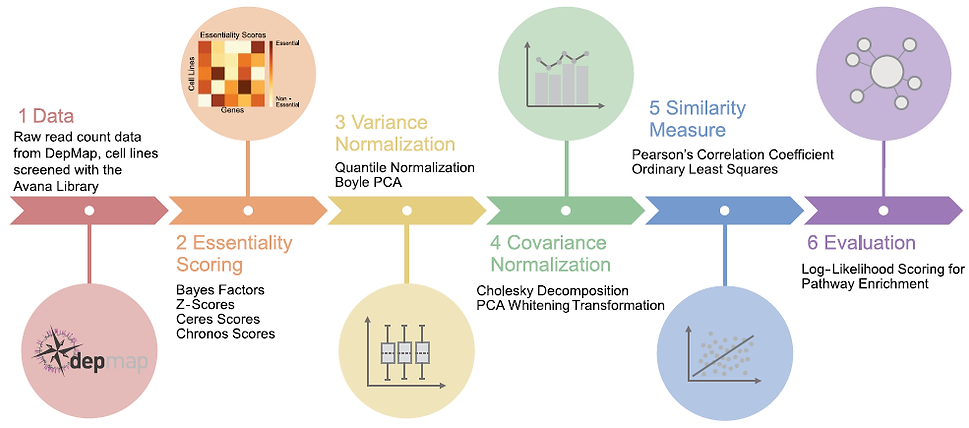
Context-Dependent Network Rewiring
Correlation networks integrate functional interactions across all cell states represented in the underlying data. But sometimes those interactions vary across cell states. This variation can give insight into cellular organization and information flow.
We use approaches like these to ask fundamental biological questions about the hierarchical organization of the cell, and to guide the development of new technologies to address the blind spots in our data.
Read the paper (Kim et al.)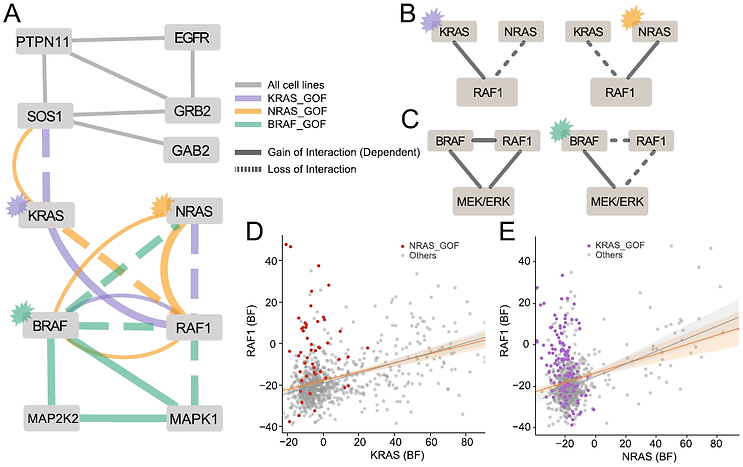
Paralog Synthetic Lethals
One major blind spot is functional buffering by paralogs, where a single gene knockout shows no phenotype in a CRISPR/Cas9 screen because a closely related gene shoulders the burden.
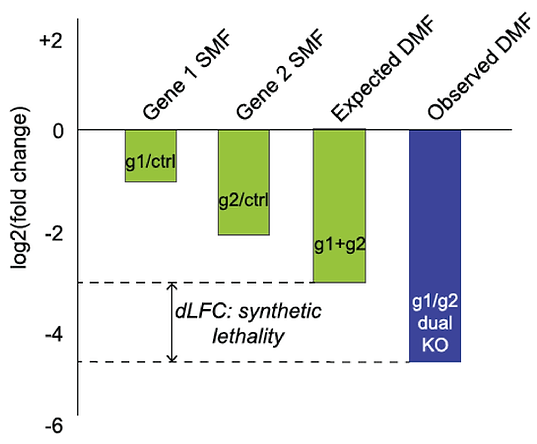
We used the enCas12a system to perform targeted double knockouts of selected paralogs. By measuring genetic interaction between gene pairs, we were able to identify dozens of synthetic lethals. For example, in the Cop9 signalosome complex, most subunits are essential but the COPS7A/B genes encode proteins that can substitute for each other.
Read the paper (Dede, McLaughlin, et al.)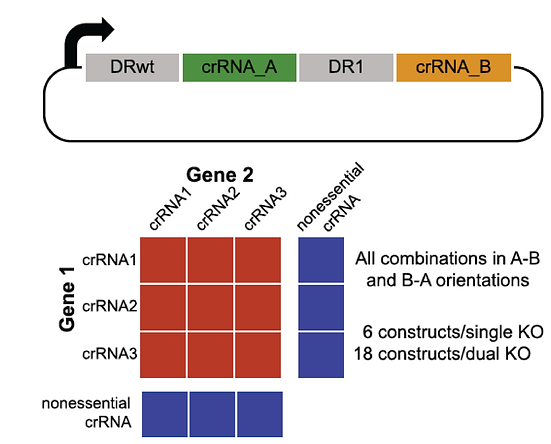
Improved Tools for Genetic Interactions
The in4mer Cas12a platform uses arrays of four independent guide RNAs targeting the same or different genes. We construct a genome-scale library, Inzolia, that is ~30% smaller than a typical CRISPR/Cas9 library while also targeting ~4000 paralog pairs. Screens in cancer cells demonstrate discrimination of core and context-dependent essential genes similar to that of CRISPR/Cas9 libraries, as well as detection of synthetic lethal and masking/buffering genetic interactions between paralogs of various family sizes. Importantly, the in4mer platform offers up to fivefold reduction in library size compared to other genetic interaction methods, substantially reducing the cost and effort required for these assays.
Read the paper (Esmaeili Anvar, Lin et al., 2024)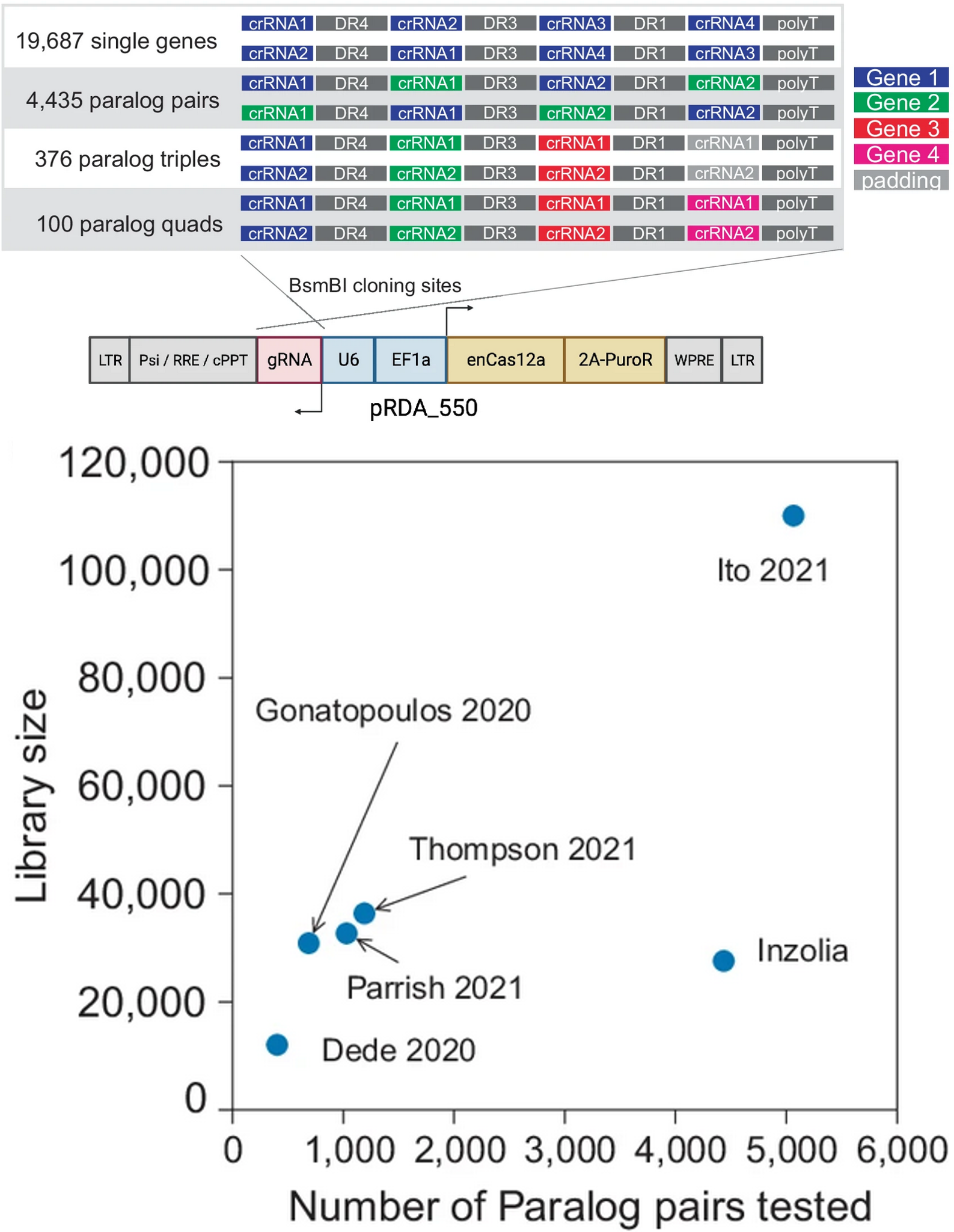
Publications
Selected peer-reviewed publications and preprints
Functional modules predict cancer-relevant genetic interactions in mammalian cells
bioRxiv. 2025; doi: https://doi.org/10.64898/2025.12.18.695201
In vivo CRISPR screens identify Mga as an immunotherapy target in triple-negative breast cancer
PNAS. 2024 Sep
FACS-based genome-wide CRISPR screens define key regulators of DNA damage signaling pathways
Molecular Cell. 2023 Aug 3;83(15):2810-2828
Genome-wide CRISPR screens using isogenic cells reveal vulnerabilities conferred by loss of tumor suppressors
Science Advances. 2022 May 13;8(19):eabm6638
Discovery of putative tumor suppressors from CRISPR screens reveals rewired lipid metabolism in acute myeloid leukemia cells
Nature Communications. 2021 Nov 11;12(1):6506
Identifying chemogenetic interactions from CRISPR screens with drugZ
Genome Medicine. 2019 Aug 12;11(1):52
Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens
G3 (Bethesda). 2017 Aug 7;7(8):2719-2727
High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities
Cell. 2015 Dec 3;163(6):1515-26
Tools
Interactive exploration of CRISPR screens
PICKLES
Explore the loss-of-function fitness profile of your favorite genes across hundreds of cell lines screened with CRISPR knockout libraries.
Launch PICKLES
Read the paper (Novak et al.)
Coessentiality Explorer
Investigate how co-essentiality and co-functionality vary by genotype and lineage.
Explore Networks
Read the paper (Kim et al.)
In4mer Design
Design your own In4mer genetic interaction screen with our library and oligo design tools.
Launch In4mer library design
Software
Open-source Python tools for screen analysis
BAGEL2 & DrugZ
Download our BAGEL2 and DrugZ software for analyzing fitness screens and drug-gene interaction screens. All software is free for any use, with attribution.
- Improved gene essentiality classification
- Multi-target correction and cross-validation
- Drug-gene interaction analysis
- Comprehensive documentation and tutorials
- Active community support
pip install drugz
# Run BAGEL2 analysis
bagel2 -i input.txt -o output.txt
# Run DrugZ analysis
drugz -i drug_screen.txt -o results/
People
Hart Lab in the Department of Systems Biology, 2025

Join Our Team
We're always looking for motivated researchers to join our interdisciplinary team. If you're interested in computational biology, CRISPR technology, or cancer research, we'd love to hear from you.